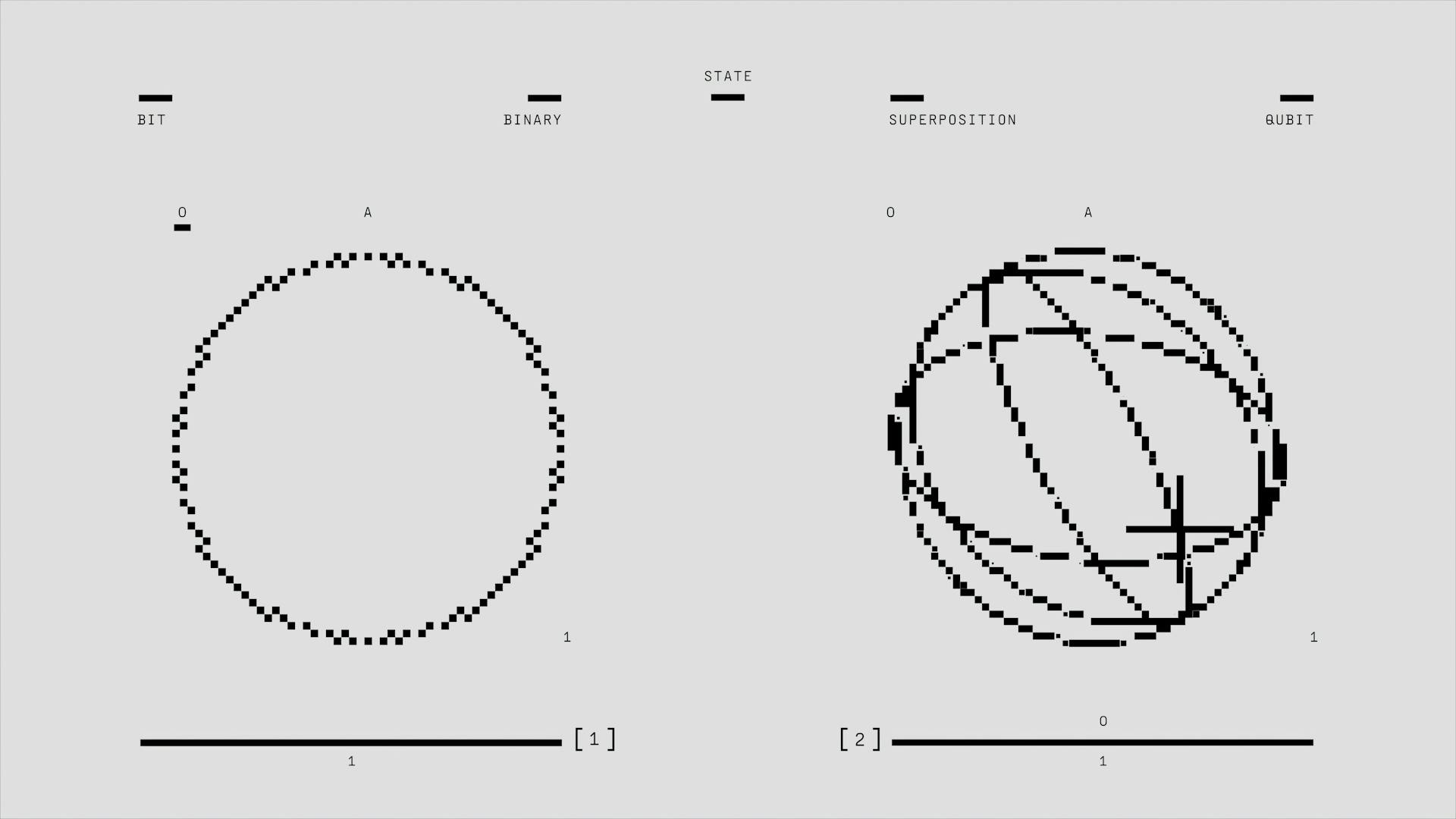
Dimension reduction is a technique used to simplify complex data by reducing the number of features or dimensions it contains. This is done to make the data easier to analyze and understand.
By reducing the number of dimensions, we can reduce the noise and irrelevant information in the data, making it more manageable. Dimension reduction helps to identify the most important features of the data.
There are several techniques used for dimension reduction, including PCA and t-SNE. PCA works by transforming the data into a new coordinate system, where the new axes are chosen to capture the most variance in the data.
Additional reading: Ai and Ml in Data Analytics
Dimension Reduction Techniques
Dimension reduction techniques are used to simplify high-dimensional data by reducing the number of variables or features while retaining the essential information. This is particularly useful when dealing with datasets that have too many variables to visualize or analyze effectively.
One way to reduce dimensions is by selecting a subset of relevant variables from the original dataset, a technique known as feature selection. This can be done by identifying the most informative variables and keeping only those.
Another approach is to find a smaller set of new variables, each being a combination of the input variables, containing the same information as the input variables. This is called dimensionality reduction, and it can be done using various techniques.
Some common dimensionality reduction techniques include linear discriminant analysis, missing value ratio, low variance filter, high correlation filter, random forest, backward feature elimination, forward feature selection, factor analysis, principal component analysis, independent component analysis, ISOMAP, t-SNE, and UMAP.
Here are some specific use cases for each technique:
By applying these techniques, we can simplify high-dimensional data and make it easier to analyze and understand.
Filtering and Selection
Filtering and selection are two important techniques used in dimensionality reduction. The filter strategy, for example, uses information gain to select relevant features.
A low variance filter is a useful technique that identifies and drops constant variables from the dataset. Variables with low variance are not useful in improving the model, and can be safely dropped. The variance of each variable should be calculated to determine which ones to drop.
Backward Feature Elimination and Forward Feature Selection are two strategies used in feature selection. However, they are time-consuming and computationally expensive, and are generally used on datasets with a small number of input variables.
Here are some common dimensionality reduction techniques:
- Low Variance filter: drop constant variables from the dataset
- High Correlation filter: drop highly correlated features
- Random Forest: find the importance of each feature and keep the topmost features
Low Variance Filter
A low variance filter is a strategy used in feature selection to identify and remove variables with low variance from a dataset. This is because variables with low variance won't affect the target variable.
Variables with low variance are essentially useless for building a model, as they won't provide any meaningful information. This is what happened with the Item_Visibility variable, which had a very low variance compared to other variables.
To apply a low variance filter, you need to calculate the variance of each variable in your dataset. This will help you identify which variables to drop and which ones to keep.
You can safely drop variables with low variance, as they won't improve your model's performance. In fact, keeping them might even hurt your model's performance.
High Correlation Filter
High Correlation Filter is a technique used to identify and remove highly correlated variables from a dataset. This can help prevent multicollinearity, where two or more variables are highly related, which can negatively impact the performance of some models.
The correlation coefficient is used to measure the strength and direction of the relationship between two variables. If the correlation coefficient crosses a certain threshold value, usually around 0.5-0.6, it's likely that the variables are highly correlated and one of them can be safely dropped.
To determine which variables to drop, you can calculate the correlation between independent numerical variables. For example, if you have a dataset with variables 'time spent on treadmill in minutes' and 'calories burnt', and the correlation coefficient is high, you can drop one of them as they likely carry similar information.
Here's a general guideline to keep in mind:
Keep in mind that this decision is highly subjective and should always be considered in the context of the domain and the specific problem you're trying to solve.
Projection Methods
Projection methods are a crucial aspect of dimension reduction. They help transform high-dimensional data into a lower-dimensional space, making it easier to analyze and visualize.
By projecting one vector onto another, dimensionality can be reduced. This is done by finding the projection of a vector onto another vector, which is essentially the vector parallel to the other vector.
Projection methods, such as PCA and LDA, can be used to extract a new set of variables from an existing large set of variables. These newly extracted variables are called Principal Components or Linear Discriminants.
Here are some key points about projection methods:
- Projection onto interesting directions
- Projection onto Manifolds
- Neighborhood Graph
- Compute Graph Distances
- Embedding
These methods can be used to reduce the dimensionality of data and make it easier to analyze.
Projection Methods
Projection methods are a class of dimensionality reduction techniques that transform data into a lower-dimensional space while preserving its essential features. This is achieved by projecting the data onto a new set of axes or dimensions.
Recommended read: Generative Ai Analytics
One common projection method is Principal Component Analysis (PCA), which uses eigenvectors to transform the data into a new space where the variance is maximized. In PCA, the first principal component explains the maximum variance in the dataset, while subsequent components explain the remaining variance.
Another projection method is Linear Discriminant Analysis (LDA), which is a generalization of Fisher's linear discriminant. LDA finds a linear combination of features that characterizes or separates two or more classes of objects or events.
Projection onto manifolds is a technique that combines multiple manifolds to represent the original data. This method involves unfolding the manifold to represent the data in a lower-dimensional space.
Uniform Manifold Approximation and Projection (UMAP) is a dimension reduction technique that preserves the local and global structure of the data. UMAP uses the concept of k-nearest neighbors and optimizes the results using stochastic gradient descent.
Here are some key advantages of UMAP:
- Handles large datasets and high-dimensional data without difficulty
- Combines the power of visualization with the ability to reduce dimensions
- Preserves both local and global structure of the data
These projection methods can be used for various applications, including data visualization, feature selection, and machine learning. By reducing the dimensionality of the data, these methods can help identify patterns and relationships that may not be apparent in the original high-dimensional space.
A unique perspective: Ai Statistical Analysis
Method 3
In the world of dimensionality reduction, there are several methods to choose from, and one of them is the t-SNE method.
The t-SNE method is a powerful technique for visualizing high-dimensional data by reducing it to a lower dimension, such as two or three. This is exactly what was done with the Fisher iris dataset, where a reducer function was generated using t-SNE.
By reducing the dimension of the iris dataset, we can gain insights into the underlying structure of the data. This is a common use case for t-SNE, where the goal is to identify patterns and relationships in the data that may not be apparent in the original high-dimensional space.
In the article, we also see an example of reducing the dimension of some images using the autoencoder method, which is another type of dimensionality reduction technique. This method is particularly useful for image data, where reducing the dimension can help to preserve the most important features of the images.
The t-SNE method is also used to generate a nonlinear data manifold, known as a Swiss-roll dataset. This dataset is a classic example of how t-SNE can be used to visualize complex data in a lower dimension.
In addition to t-SNE, other methods like isomap and locally linear embedding (LLE) can also be used for dimensionality reduction. These methods have their own strengths and weaknesses, and the choice of method depends on the specific characteristics of the data.
In terms of performance, the t-SNE method can be quite computationally intensive, especially for large datasets. However, the article notes that the timing for the t-SNE method can be compared to the default CPU computation, which can be a useful benchmark for evaluating the performance of different methods.
Feature Selection and Elimination
Feature selection is a crucial step in dimension reduction. It involves selecting a subset of the most relevant features that contribute to the model's performance.
There are three strategies for feature selection: the filter strategy (e.g., information gain), the wrapper strategy (e.g., accuracy-guided search), and the embedded strategy (features are added or removed while building the model based on prediction errors). Data analysis such as regression or classification can be done in the reduced space more accurately than in the original space.
Two popular techniques for feature selection are Backward Feature Elimination and Forward Feature Selection. Backward Feature Elimination involves eliminating one feature at a time, while Forward Feature Selection involves selecting the best features that enhance the model's performance.
Here are the steps to implement Backward Feature Elimination:
- We first take all the n variables present in our dataset and train the model using them
- We then calculate the performance of the model
- Now, we compute the performance of the model after eliminating each variable (n times), i.e., we drop one variable every time and train the model on the remaining n-1 variables
- We identify the variable whose removal has produced the smallest (or no) change in the performance of the model and then drop that variable
- Repeat this process until no variable can be dropped
Note that both Backward Feature Elimination and Forward Feature Selection are time-consuming and computationally expensive. They are practically only used on datasets with a small number of input variables.
Suggestion: What Is Feature Engineering in Data Science
NMF
NMF is a powerful tool in fields where only non-negative signals exist, such as astronomy.
It decomposes a non-negative matrix into the product of two non-negative ones, which has been a promising approach in various applications.
The multiplicative update rule by Lee & Seung is well known for its continuous development, including the inclusion of uncertainties and handling missing data.
Sequential NMF preserves the flux in direct imaging of circumstellar structures in astronomy, making it useful for detecting exoplanets.
It's able to preserve more information than PCA by not removing the mean of the matrices, resulting in physical non-negative fluxes.
NMF has been continuously developed to handle missing data and parallel computation, making it a stable and efficient method.
Feature Selection
Feature selection is a crucial step in data analysis that aims to find a suitable subset of input variables for the task at hand. This process can be done using various strategies, including the filter strategy (e.g., information gain), the wrapper strategy (e.g., accuracy-guided search), and the embedded strategy (features are added or removed while building the model based on prediction errors).
There are three main strategies for feature selection: filter, wrapper, and embedded. The filter strategy is used for feature selection before building a model, the wrapper strategy is used for feature selection while building a model, and the embedded strategy is used for feature selection during model building.
Here's an interesting read: Feature Engineering Python
Backward feature elimination is a technique that eliminates features one by one based on their performance impact on the model. This process starts with all the variables present in the dataset and trains the model using them. The variable whose removal has produced the smallest (or no) change in the model's performance is then dropped.
Forward feature selection is a technique that adds features one by one based on their performance impact on the model. This process starts with a single feature and trains the model n number of times using each feature separately. The variable that produces the highest increase in performance is then retained.
Random Forest is a widely used algorithm for feature selection that comes packaged with in-built feature importance. This helps us select a smaller subset of features without needing to program feature importance separately.
Here are some popular feature selection techniques:
- Backward Feature Elimination
- Forward Feature Selection
- Random Forest
- UMAP (Uniform manifold approximation and projection)
These techniques can be used to reduce the dimensionality of the data and improve the performance of the model.
Independent Component
Independent components are a crucial part of dimensionality reduction techniques, and one of the most widely used methods is Independent Component Analysis (ICA). ICA is based on information theory and looks for independent factors, not just uncorrelated ones.
The key difference between ICA and PCA is that PCA looks for uncorrelated factors, while ICA looks for independent factors. This means that if two variables are independent, they are not dependent on other variables.
ICA assumes that the given variables are linear mixtures of some unknown latent variables, which are mutually independent. These latent variables are called the independent components of the observed data.
To find the independent components, we need to find an un-mixing matrix that makes the components as independent as possible. Non-Gaussianity is a common method to measure the independence of components.
The distribution of the sum of independent components tends to be normally distributed (Gaussian), so we look for transformations that maximize the kurtosis of each component. Kurtosis is the third-order moment of the distribution, and maximizing it makes the distribution non-gaussian, which in turn makes the components independent.
Here's a summary of the key points about independent components:
- Independent components are mutually independent and not dependent on other variables.
- ICA looks for independent factors, not just uncorrelated ones.
- Non-Gaussianity is used to measure the independence of components.
- Kurtosis is used to maximize the independence of components.
Frequently Asked Questions
When would you reduce dimensions in your data?
You would reduce dimensions in your data when dealing with large datasets to lower computation costs or when your model is underperforming due to an ill-fitting feature set. This can help improve model accuracy and efficiency.
What is dimensionality reduction in PCA?
Dimensionality reduction in PCA involves transforming a large set of variables into a smaller one that retains most of the original information. This process helps to simplify complex data while preserving its essential characteristics.
What is data reduction in machine learning?
Data reduction is a method that shrinks large datasets to a smaller size while preserving their integrity, making them more manageable for analysis. By reducing data size, you can save time in the long run, but don't overlook the time spent on data reduction itself.
Sources
- https://en.wikipedia.org/wiki/Dimensionality_reduction
- https://www.geeksforgeeks.org/dimensionality-reduction/
- https://www.analyticsvidhya.com/blog/2018/08/dimensionality-reduction-techniques-python/
- https://reference.wolfram.com/language/ref/DimensionReduction.html.en
- https://en.wikipedia.org/wiki/Dimensional_reduction
Featured Images: pexels.com